Manipulate PMMH objects.
Source:R/pmmh.r
Manipulate PMMH objects.
The method biips_pmmh_update performs adaptation and burn-in
iterations for the PMMH algorithm.
The method biips_pmmh_samples performs iterations for the PMMH
algorithm and returns samples.
is.pmmh(object) # S3 method for pmmh biips_pmmh_update(object, n_iter, n_part, thin = 1, max_fail = 0, rw_adapt = TRUE, output = "p", ...) # S3 method for pmmh biips_pmmh_samples(object, n_iter, n_part, thin = 1, max_fail = 0, output = "p", ...)
Arguments
| object | a |
|---|---|
| n_iter | integer. Number of burn-in iterations. |
| n_part | integer. Number of particles used in SMC algorithms. |
| thin | integer. Thinning interval. Returns samples every |
| max_fail | integer. maximum number of failed SMC algorithms allowed. (default=0). |
| rw_adapt | logical. Activate adaptation of the proposal
(default= |
| output | string. Select additional members to be returned in the
|
| ... | Additional arguments to be passed to the SMC
algorithm such as |
Value
The function is.pmmh returns TRUE if the object is of
class pmmh.
The methods biips_pmmh_update and biips_pmmh_update
return an object of class mcmcarray.list.
biips_pmmh_samples output contains one mcmcarray
member for each monitored variable returned by the param_names() and
latent_names() member functions of the pmmh object.
The members of the mcmcarray.list object are
mcmcarray objects for different variables. Assuming
dim is the dimension of the monitored variable, the
mcmcarray object is an array of dimension c(dim,
n_iter) with the following attributes (accessible with
attr):
string with the name of the variable.
vector with the lower bounds of the variable.
vector with the upper bounds of the variable.
Details
The output string arguments can be used to query additional
members in the mcmcarray.list output. If output
contains:
- p
returns member
log_marg_like_pen.mcmcarraywith penalized log marginal likelihood estimates over iterations.- l
returns member
log_marg_like.mcmcarraywith log marginal likelihood estimates over iterations.- a
returns member
info$accept_rate.mcmcarraywith acceptance rate over iterations.- s
returns member
info$rw_step.mcmcarraywith standard deviations of the random walk over iterations.- f
returns member
info$n_fail. number of failed SMC algorithms.
See also
Examples
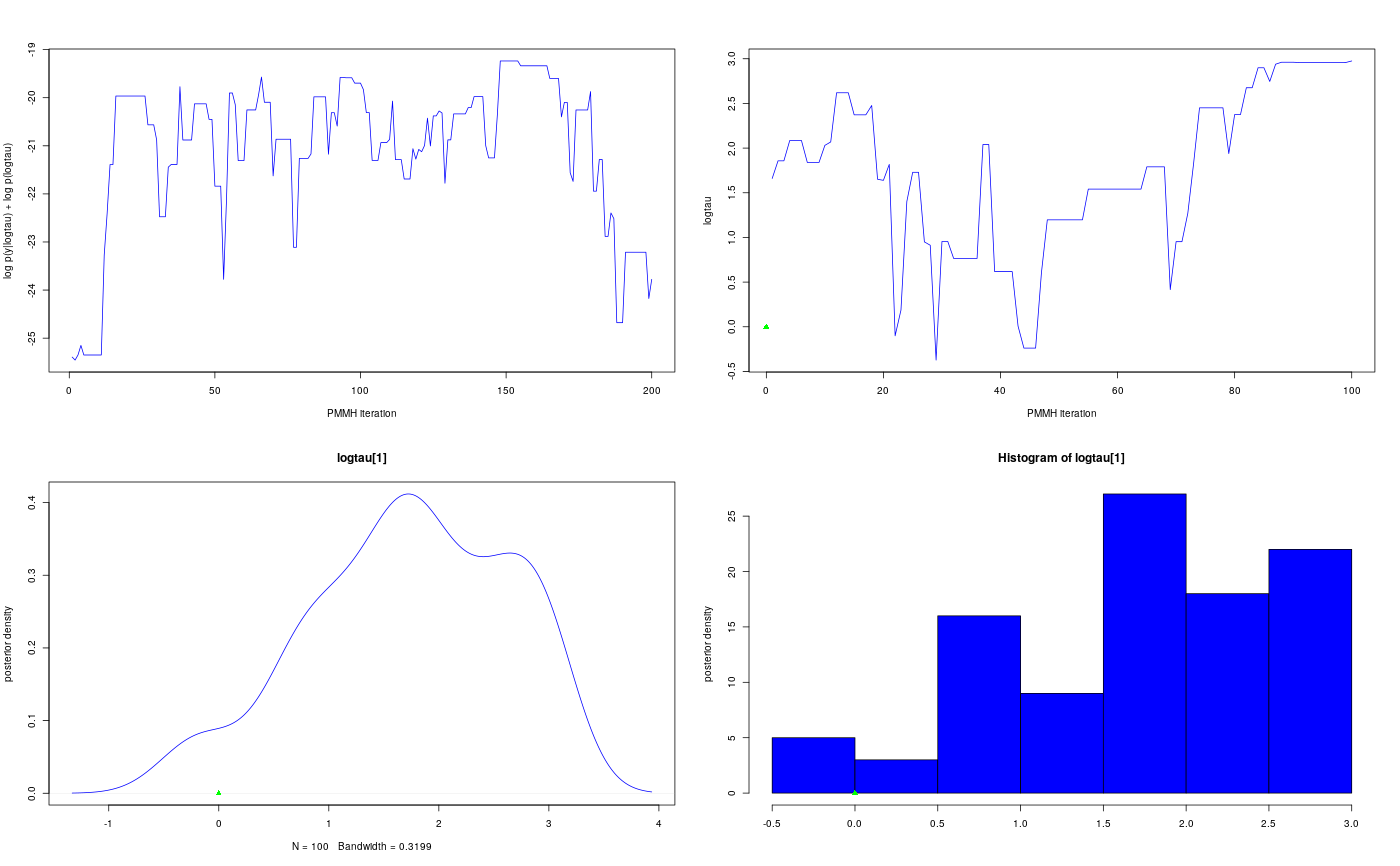
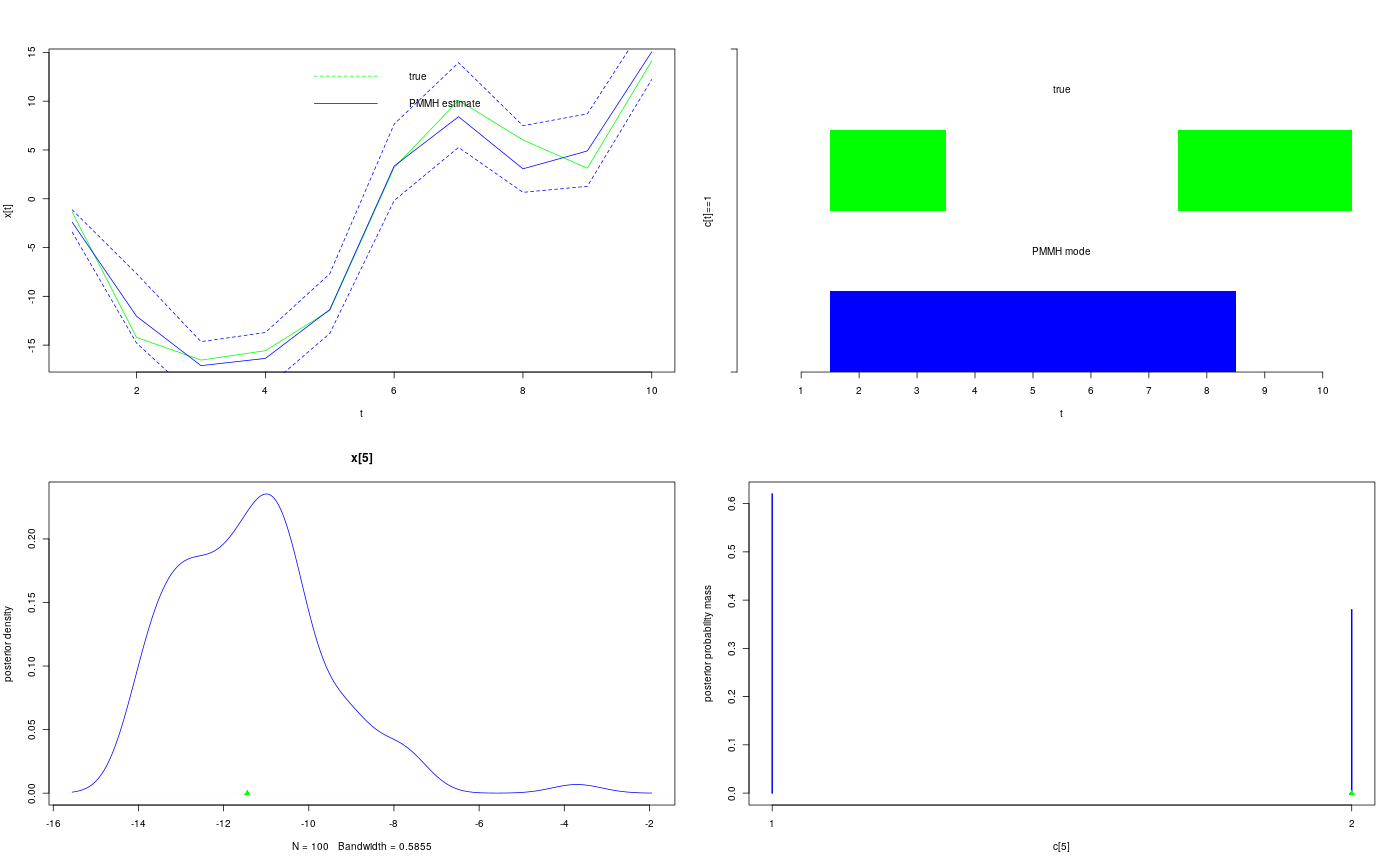
modelfile <- system.file('extdata', 'hmm.bug', package = 'rbiips') stopifnot(nchar(modelfile) > 0) cat(readLines(modelfile), sep = '\n')#> var c_true[tmax], x_true[tmax], c[tmax], x[tmax], y[tmax] #> #> data #> { #> x_true[1] ~ dnorm(0, 1/5) #> y[1] ~ dnorm(x_true[1], exp(logtau_true)) #> for (t in 2:tmax) #> { #> c_true[t] ~ dcat(p) #> x_true[t] ~ dnorm(0.5*x_true[t-1]+25*x_true[t-1]/(1+x_true[t-1]^2)+8*cos(1.2*(t-1)), ifelse(c_true[t]==1, 1/10, 1/100)) #> y[t] ~ dnorm(x_true[t]/4, exp(logtau_true)) #> } #> } #> #> model #> { #> logtau ~ dunif(-3, 3) #> x[1] ~ dnorm(0, 1/5) #> y[1] ~ dnorm(x[1], exp(logtau)) #> for (t in 2:tmax) #> { #> c[t] ~ dcat(p) #> x[t] ~ dnorm(0.5*x[t-1]+25*x[t-1]/(1+x[t-1]^2)+8*cos(1.2*(t-1)), ifelse(c[t]==1, 1/10, 1/100)) #> y[t] ~ dnorm(x[t]/4, exp(logtau)) #> } #> }#> * Parsing model in: /home/adrien/Dropbox/workspace/rbiips/inst/extdata/hmm.bug #> * Compiling data graph #> Declaring variables #> Resolving undeclared variables #> Allocating nodes #> Graph size: 168 #> Sampling data #> Reading data back into data table #> * Compiling model graph #> Declaring variables #> Resolving undeclared variables #> Allocating nodes #> Graph size: 180n_part <- 50 obj_pmmh <- biips_pmmh_init(model, 'logtau', latent_names = c('x', 'c[2:10]'), inits = list(logtau = -2)) # Initialize#> * Initializing PMMHis.pmmh(obj_pmmh)#> [1] TRUEout_pmmh_burn <- biips_pmmh_update(obj_pmmh, 100, n_part) # Burn-in#> * Adapting PMMH with 50 particles #> |--------------------------------------------------| 100% #> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100 iterations in 0.29 sout_pmmh <- biips_pmmh_samples(obj_pmmh, 100, n_part, thin = 1) # Samples#> * Generating 100 PMMH samples with 50 particles #> |--------------------------------------------------| 100% #> |**************************************************| 100 iterations in 0.28 sdens_pmmh_lt <- biips_density(out_pmmh$logtau) summ_pmmh_x <- biips_summary(out_pmmh$x, order = 2, probs = c(0.025, 0.975)) dens_pmmh_x <- biips_density(out_pmmh$x) summ_pmmh_c <- biips_summary(out_pmmh[['c[2:10]']]) table_pmmh_c <- biips_table(out_pmmh[['c[2:10]']]) par(mfrow = c(2, 2)) plot(c(out_pmmh_burn$log_marg_like_pen, out_pmmh$log_marg_like_pen), type = 'l', col = 'blue', xlab = 'PMMH iteration', ylab = 'log p(y|logtau) + log p(logtau)') plot(out_pmmh$logtau[1, ], type = 'l', col = 'blue', xlab = 'PMMH iteration', ylab = 'logtau') points(0, model$data()$logtau_true, pch = 17, col = 'green') plot(dens_pmmh_lt, col = 'blue', ylab = 'posterior density') points(model$data()$logtau_true, 0, pch = 17, col = 'green') biips_hist(out_pmmh$logtau, col = 'blue', ylab = 'posterior density')points(model$data()$logtau_true, 0, pch = 17, col = 'green')par(mfrow = c(2, 2)) plot(model$data()$x_true, type = 'l', col = 'green', xlab = 't', ylab = 'x[t]') lines(summ_pmmh_x$mean, col = 'blue') matlines(matrix(unlist(summ_pmmh_x$quant), data$tmax), lty = 2, col = 'blue') legend('topright', leg = c('true', 'PMMH estimate'), lty = c(2, 1), col = c('green', 'blue'), bty = 'n') barplot(.5*(model$data()$c_true==1), col = 'green', border = NA, space = 0, offset = 1, ylim=c(0,2), xlab='t', ylab='c[t]==1', axes = FALSE) axis(1, at=1:data$tmax-.5, labels=1:data$tmax) axis(2, line = 1, at=c(0,2), labels=NA) text(data$tmax/2, 1.75, 'true') barplot(.5*c(NA, summ_pmmh_c$mode==1), col = 'blue', border = NA, space = 0, axes = FALSE, add = TRUE) text(data$tmax/2, .75, 'PMMH mode') t <- 5 plot(dens_pmmh_x[[t]], col='blue', ylab = 'posterior density') points(model$data()$x_true[t], 0, pch = 17, col = 'green') plot(table_pmmh_c[[t-1]], col='blue', ylab = 'posterior probability mass')points(model$data()$c_true[t], 0, pch = 17, col = 'green')