Matbiips: Tutorial 2
In this tutorial, we consider applying sequential Monte Carlo methods for sensitivity analysis and parameter estimation in a nonlinear non-Gaussian hidden Markov model.
Contents
Statistical model
The statistical model is defined as follows.
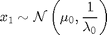
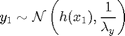
For 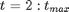
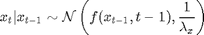
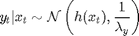
where 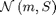 denotes the Gaussian distribution of mean
denotes the Gaussian distribution of mean  and covariance matrix
and covariance matrix 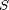 ,
, 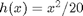 ,
, 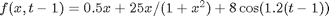 ,
, 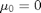 ,
, 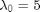 ,
, 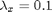 . The precision of the observation noise
. The precision of the observation noise 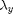 is also assumed to be unknown. We will assume a uniform prior for
is also assumed to be unknown. We will assume a uniform prior for 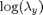 :
:
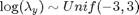
Statistical model in BUGS language
We describe the model in BUGS language in the file 'hmm_1d_nonlin.bug':
model_file = 'hmm_1d_nonlin_param.bug'; % BUGS model filename type(model_file);
var x_true[t_max], x[t_max], y[t_max]
data
{
#log_prec_y_true ~ dunif(-3, 3)
prec_y_true <- exp(log_prec_y_true)
x_true[1] ~ dnorm(mean_x_init, prec_x_init)
y[1] ~ dnorm(x_true[1]^2/20, prec_y_true)
for (t in 2:t_max)
{
x_true[t] ~ dnorm(0.5*x_true[t-1]+25*x_true[t-1]/(1+x_true[t-1]^2)+8*cos(1.2*(t-1)), prec_x)
y[t] ~ dnorm(x_true[t]^2/20, prec_y_true)
}
}
model
{
log_prec_y ~ dunif(-3, 3)
prec_y <- exp(log_prec_y)
x[1] ~ dnorm(mean_x_init, prec_x_init)
y[1] ~ dnorm(x[1]^2/20, prec_y)
for (t in 2:t_max)
{
x[t] ~ dnorm(0.5*x[t-1]+25*x[t-1]/(1+x[t-1]^2)+8*cos(1.2*(t-1)), prec_x)
y[t] ~ dnorm(x[t]^2/20, prec_y)
}
}
Installation of Matbiips
- Download the latest version of Matbiips
- Unzip the archive in some folder
- Add the Matbiips folder to the Matlab search path
matbiips_path = '../../matbiips';
addpath(matbiips_path)
General settings
set(0, 'DefaultAxesFontsize', 14); set(0, 'Defaultlinelinewidth', 2); light_blue = [.7, .7, 1];
Set the random numbers generator seed for reproducibility
if isoctave() || verLessThan('matlab', '7.12') rand('state', 0) else rng('default') end
Load model and data
Model parameters
t_max = 20; mean_x_init = 0; prec_x_init = 1; prec_x = 10; log_prec_y_true = log(1); % True value used to sample the data data = struct('t_max', t_max, 'prec_x_init', prec_x_init,... 'prec_x', prec_x, 'log_prec_y_true', log_prec_y_true,... 'mean_x_init', mean_x_init);
Compile BUGS model and sample data
sample_data = true; % Boolean model = biips_model(model_file, data, 'sample_data', sample_data); % Create Biips model and sample data data = model.data;
* Parsing model in: hmm_1d_nonlin_param.bug * Compiling data graph Declaring variables Resolving undeclared variables Allocating nodes Graph size: 280 Sampling data Reading data back into data table * Compiling model graph Declaring variables Resolving undeclared variables Allocating nodes Graph size: 284
Biips Sensitivity analysis with Sequential Monte Carlo
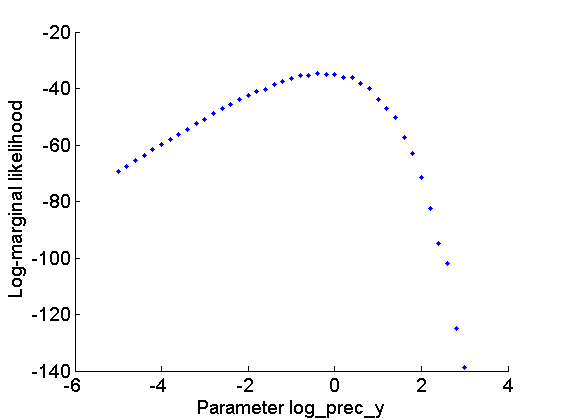
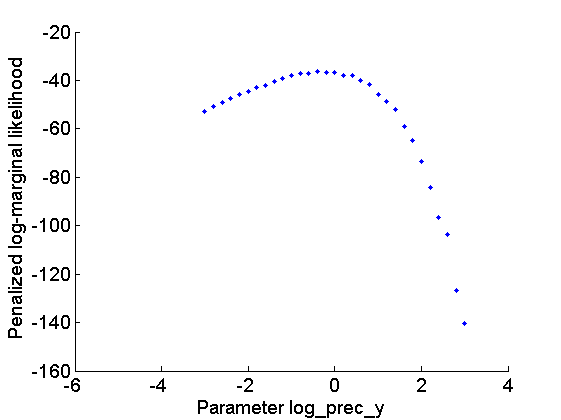
Let now use Biips to provide estimates of the marginal log-likelihood and penalized marginal log-likelihood given various values of the log-precision parameters 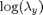 .
.
Parameters of the algorithm.
n_part = 100; % Number of particles param_names = {'log_prec_y'}; % Parameter for which we want to study sensitivity param_values = {-5:.2:3}; % Range of values
Run sensitivity analysis with SMC
out_sens = biips_smc_sensitivity(model, param_names, param_values, n_part);
* Analyzing sensitivity with 100 particles |--------------------------------------------------| 100% |**************************************************| 41 iterations in 1.27 s
Plot log-marginal likelihood and penalized log-marginal likelihood
figure('name', 'Log-marginal likelihood'); plot(param_values{1}, out_sens.log_marg_like, '.') xlabel('Parameter log\_prec\_y') ylabel('Log-marginal likelihood') box off figure('name', 'Penalized log-marginal likelihood'); plot(param_values{1}, out_sens.log_marg_like_pen, '.') xlabel('Parameter log\_prec\_y') ylabel('Penalized log-marginal likelihood') box off
Biips Particle Marginal Metropolis-Hastings
We now use Biips to run a Particle Marginal Metropolis-Hastings in order to obtain posterior MCMC samples of the parameter and the variables  .
.
Parameters of the PMMH.
param_names indicates the parameters to be sampled using a random walk Metroplis-Hastings step. For all the other variables, Biips will use a sequential Monte Carlo as proposal.
n_burn = 2000; % nb of burn-in/adaptation iterations n_iter = 2000; % nb of iterations after burn-in thin = 1; % thinning of MCMC outputs n_part = 50; % nb of particles for the SMC var_name = 'log_prec_y'; param_names = {var_name}; % name of the variables updated with MCMC (others are updated with SMC) latent_names = {'x'}; % name of the variables updated with SMC and that need to be monitored
Init PMMH
obj_pmmh = biips_pmmh_init(model, param_names, 'inits', {-2},... 'latent_names', latent_names); % creates a pmmh object
* Initializing PMMH
Run PMMH
obj_pmmh = biips_pmmh_update(obj_pmmh, n_burn, n_part); % adaptation and burn-in iterations [obj_pmmh, out_pmmh, log_marg_like_pen, log_marg_like, stats_pmmh] = biips_pmmh_samples(obj_pmmh, n_iter, n_part,... 'thin', thin); % samples
* Adapting PMMH with 50 particles |--------------------------------------------------| 100% |++++++++++++++++++++++++++++++++++++++++++++++++++| 2000 iterations in 15.14 s * Generating 2000 PMMH samples with 50 particles |--------------------------------------------------| 100% |**************************************************| 2000 iterations in 14.09 s
Some summary statistics
summ_pmmh = biips_summary(out_pmmh, 'probs', [.025, .975]);
Compute kernel density estimates
kde_pmmh = biips_density(out_pmmh);
Posterior mean and credible interval of the parameter
summ_param = getfield(summ_pmmh, var_name); fprintf('Posterior mean of %s: %.1f\n', var_name, summ_param.mean); fprintf('95%% credible interval of %s: [%.1f, %.1f]\n', var_name, ... summ_param.quant{1}, summ_param.quant{2});
Posterior mean of log_prec_y: -0.4 95% credible interval of log_prec_y: [-1.2, 0.4]
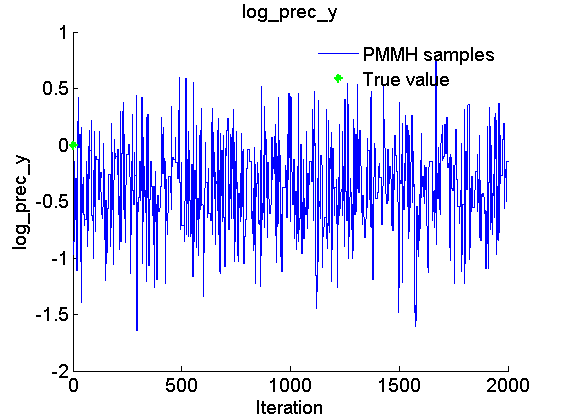
Trace of MCMC samples for the parameter
figure('name', 'PMMH: Trace samples parameter') samples_param = getfield(out_pmmh, var_name); param_lab = 'log\_prec\_y'; plot(samples_param, 'linewidth', 1) hold on plot(0, data.log_prec_y_true, '*g'); xlabel('Iteration') ylabel(param_lab) title(param_lab) legend({'PMMH samples', 'True value'}) legend boxoff box off
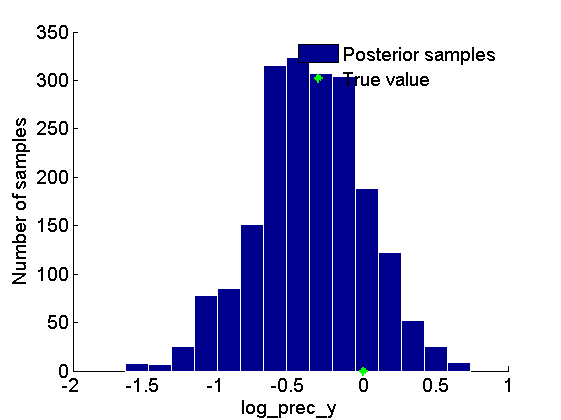
Histogram and kde estimate of the posterior for the parameter
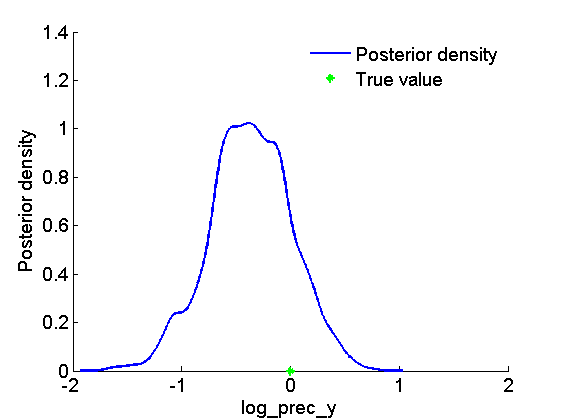
figure('name', 'PMMH: Histogram posterior parameter') hist(samples_param, 15) h = findobj(gca, 'Type', 'patch'); set(h, 'EdgeColor', 'w') hold on plot(data.log_prec_y_true, 0, '*g'); xlabel(param_lab) ylabel('Number of samples') legend({'Posterior samples', 'True value'}) legend boxoff box off figure('name', 'PMMH: KDE estimate posterior parameter') kde_param = getfield(kde_pmmh, var_name); plot(kde_param.x, kde_param.f); hold on plot(data.log_prec_y_true, 0, '*g'); xlabel(param_lab); ylabel('Posterior density'); legend({'Posterior density', 'True value'}) legend boxoff box off
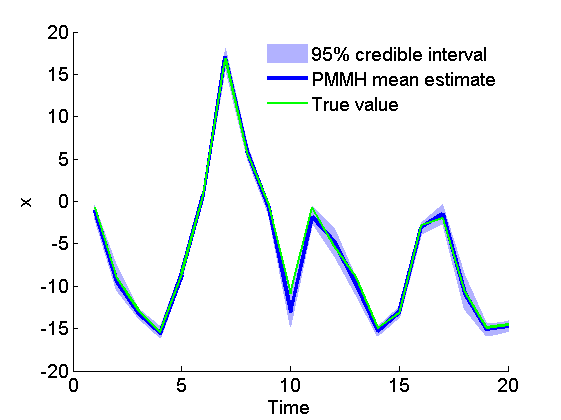
Posterior mean and quantiles for x
figure('name', 'PMMH: Posterior mean and quantiles') x_pmmh_mean = summ_pmmh.x.mean; x_pmmh_quant = summ_pmmh.x.quant; h = fill([1:t_max, t_max:-1:1], [x_pmmh_quant{1}; flipud(x_pmmh_quant{2})], 0); set(h, 'edgecolor', 'none', 'facecolor', light_blue) hold on plot(1:t_max, x_pmmh_mean, 'linewidth', 3) plot(1:t_max, data.x_true, 'g') xlabel('Time') ylabel('x') legend({'95% credible interval', 'PMMH mean estimate', 'True value'}) box off legend boxoff
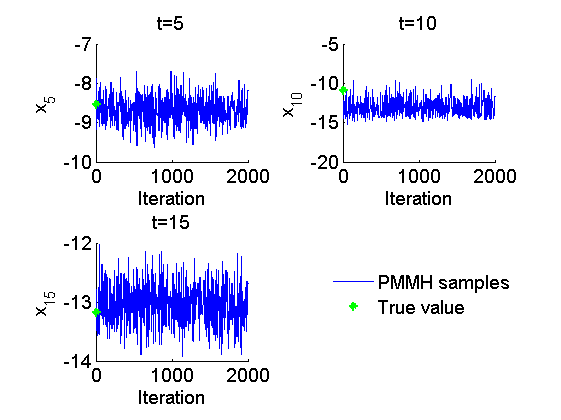
Trace of MCMC samples for x
figure('name', 'PMMH: Trace samples x') time_index = [5, 10, 15]; for k=1:numel(time_index) tk = time_index(k); subplot(2, 2, k) plot(out_pmmh.x(tk, :), 'linewidth', 1) hold on plot(0, data.x_true(tk), '*g'); xlabel('Iteration') ylabel(['x_{', num2str(tk), '}']) title(['t=', num2str(tk)]); box off end h = legend({'PMMH samples', 'True value'}); set(h, 'position', [0.7, 0.25, .1, .1]) legend boxoff
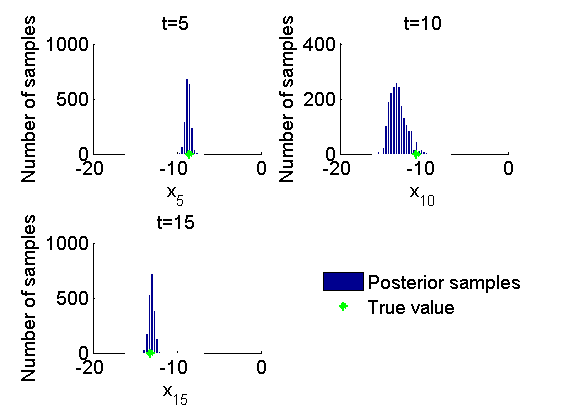
Histogram and kernel density estimate of posteriors of x
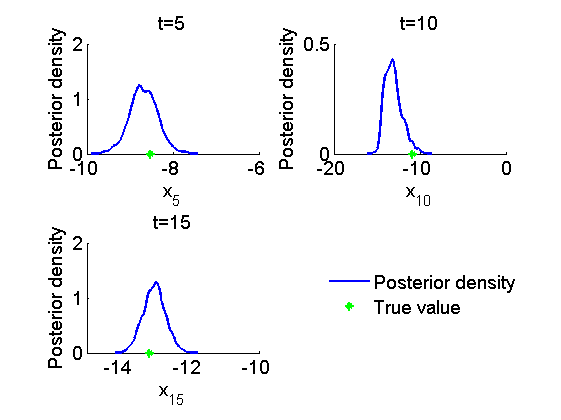
figure('name', 'PMMH: Histograms marginal posteriors') for k=1:numel(time_index) tk = time_index(k); subplot(2, 2, k) hist(out_pmmh.x(tk, :), -16:.3:-7); h = findobj(gca, 'Type', 'patch'); set(h, 'EdgeColor', 'w') hold on plot(data.x_true(tk), 0, '*g'); xlabel(['x_{', num2str(tk), '}']); ylabel('Number of samples'); title(['t=', num2str(tk)]); box off end h = legend({'Posterior samples', 'True value'}); set(h, 'position', [0.7, 0.25, .1, .1]) legend boxoff figure('name', 'PMMH: KDE estimates marginal posteriors') for k=1:numel(time_index) tk = time_index(k); subplot(2, 2, k) plot(kde_pmmh.x(tk).x, kde_pmmh.x(tk).f); hold on plot(data.x_true(tk), 0, '*g'); xlabel(['x_{', num2str(tk), '}']); ylabel('Posterior density'); title(['t=', num2str(tk)]); box off end h = legend({'Posterior density', 'True value'}); set(h, 'position', [0.7, 0.25, .1, .1]); legend boxoff
Clear model
biips_clear()