Matbiips example: Object tracking
In this example, we consider the tracking of an object in 2D, observed by a radar.
Reference: B. Ristic. Beyond the Kalman filter: Particle filters for Tracking applications. Artech House, 2004.
Contents
Statistical model
Let 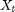 be a 4-D vector containing the position and velocity of an object in 2D. We obtain distance-angular measurements
be a 4-D vector containing the position and velocity of an object in 2D. We obtain distance-angular measurements 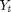 from a radar.
from a radar.
The model is defined as follows. For 
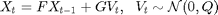
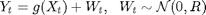
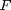 and
and 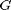 are known matrices,
are known matrices, 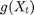 is the known nonlinear measurement function and
is the known nonlinear measurement function and 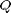 and
and 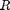 are known covariance matrices.
are known covariance matrices.
Statistical model in BUGS language
model_file = 'hmm_4d_nonlin_tracking.bug'; % BUGS model filename type(model_file);
var v_true[2,t_max-1], x_true[4,t_max], x_radar_true[2,t_max],
v[2,t_max-1], x[4,t_max], x_radar[2,t_max], y[2,t_max]
data
{
x_true[,1] ~ dmnorm(mean_x_init, prec_x_init)
x_radar_true[,1] <- x_true[1:2,1] - x_pos
mu_y_true[1,1] <- sqrt(x_radar_true[1,1]^2+x_radar_true[2,1]^2)
mu_y_true[2,1] <- arctan(x_radar_true[2,1]/x_radar_true[1,1])
y[,1] ~ dmnorm(mu_y_true[,1], prec_y)
for (t in 2:t_max)
{
v_true[,t-1] ~ dmnorm(mean_v, prec_v)
x_true[,t] <- F %*% x_true[,t-1] + G %*% v_true[,t-1]
x_radar_true[,t] <- x_true[1:2,t] - x_pos
mu_y_true[1,t] <- sqrt(x_radar_true[1,t]^2+x_radar_true[2,t]^2)
mu_y_true[2,t] <- arctan(x_radar_true[2,t]/x_radar_true[1,t])
y[,t] ~ dmnorm(mu_y_true[,t], prec_y)
}
}
model
{
x[,1] ~ dmnorm(mean_x_init, prec_x_init)
x_radar[,1] <- x[1:2,1] - x_pos
mu_y[1,1] <- sqrt(x_radar[1,1]^2+x_radar[2,1]^2)
mu_y[2,1] <- arctan(x_radar[2,1]/x_radar[1,1])
y[,1] ~ dmnorm(mu_y[,1], prec_y)
for (t in 2:t_max)
{
v[,t-1] ~ dmnorm(mean_v, prec_v)
x[,t] <- F %*% x[,t-1] + G %*% v[,t-1]
x_radar[,t] <- x[1:2,t] - x_pos
mu_y[1,t] <- sqrt(x_radar[1,t]^2+x_radar[2,t]^2)
mu_y[2,t] <- arctan(x_radar[2,t]/x_radar[1,t])
y[,t] ~ dmnorm(mu_y[,t], prec_y)
}
}
Installation of Matbiips
- Download the latest version of Matbiips
- Unzip the archive in some folder
- Add the Matbiips folder to the Matlab search path
matbiips_path = '../../matbiips';
addpath(matbiips_path)
General settings
set(0, 'DefaultAxesFontsize', 14); set(0, 'Defaultlinelinewidth', 2); light_blue = [.7, .7, 1]; light_red = [1, .7, .7]; % Set the random numbers generator seed for reproducibility if isoctave() || verLessThan('matlab', '7.12') rand('state', 0) else rng('default') end
Load model and data
Model parameters
t_max = 20;
mean_x_init = [0, 0, 1, 0]';
prec_x_init = 1000*eye(4);
x_pos = [60, 0];
mean_v = zeros(2, 1);
prec_v = eye(2);
prec_y = diag([100 500]);
delta_t = 1;
F =[1, 0, delta_t, 0;
0, 1, 0, delta_t;
0, 0, 1, 0;
0, 0, 0, 1];
G = [delta_t.^2/2, 0;
0, delta_t.^2/2;
delta_t, 0;
0, delta_t];
data = struct('t_max', t_max, 'mean_x_init', mean_x_init, 'prec_x_init', ...
prec_x_init, 'x_pos', x_pos, 'mean_v', mean_v, 'prec_v', prec_v,...
'prec_y', prec_y, 'F', F, 'G', G);
Compile BUGS model and sample data
sample_data = true; % Boolean model = biips_model(model_file, data, 'sample_data', sample_data); data = model.data; x_pos_true = data.x_true(1:2,:);
* Parsing model in: hmm_4d_nonlin_tracking.bug * Compiling data graph Declaring variables Resolving undeclared variables Allocating nodes Graph size: 327 Sampling data Reading data back into data table * Compiling model graph Declaring variables Resolving undeclared variables Allocating nodes Graph size: 331
Biips Particle filter
Parameters of the algorithm
n_part = 100000; % Number of particles variables = {'x'}; % Variables to be monitored
Run SMC
out_smc = biips_smc_samples(model, {'x'}, n_part);
* Assigning node samplers * Running SMC forward sampler with 100000 particles |--------------------------------------------------| 100% |**************************************************| 20 iterations in 45.73 s
Diagnostic
diag_smc = biips_diagnosis(out_smc);
* Diagnosis of variable: x[1:4,1:20] Filtering: GOOD Smoothing: GOOD
Summary statistics
summ_smc = biips_summary(out_smc, 'probs', [.025, .975]);
Plot estimates
figure('name', 'SMC: Filtering and smoothing estimates') x_f_mean = summ_smc.x.f.mean; x_s_mean = summ_smc.x.s.mean; plot(x_f_mean(1, :), x_f_mean(2, :)) hold on plot(x_s_mean(1, :), x_s_mean(2, :), '-.r') plot(x_pos_true(1,:), x_pos_true(2,:), '--g') plot(x_pos(1), x_pos(2), 'sk') legend('Filtering estimate', 'Smoothing estimate', 'True trajectory',... 'Position of the radar', 'location', 'Northwest') legend boxoff xlabel('Position X') ylabel('Position Y') box off
Plot particles
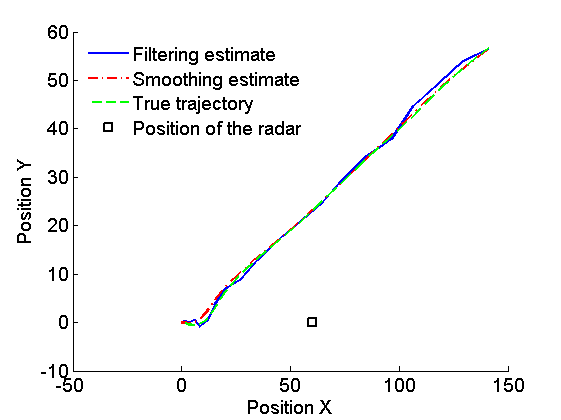
figure('name', 'SMC: Particles (filtering)') plot(out_smc.x.f.values(1,:), out_smc.x.f.values(2,:), 'ro', ... 'markersize', 2, 'markerfacecolor', 'r') hold on plot(x_pos_true(1,:), x_pos_true(2,:), '--g') plot(x_pos(1), x_pos(2), 'sk') legend('Particles (filtering)', 'True trajectory', 'Position of the radar', 'location', 'Northwest') legend boxoff xlabel('Position X') ylabel('Position Y') box off
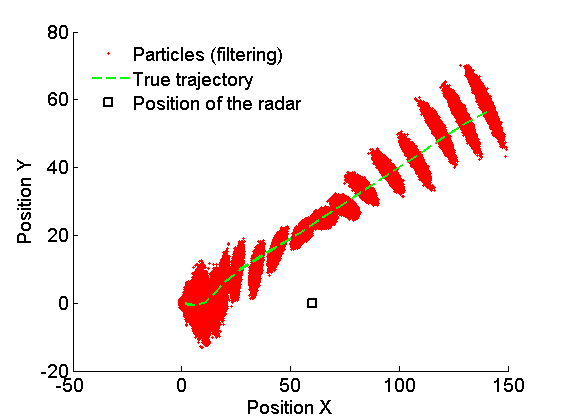
Plot Filtering estimates
x_f_quant = summ_smc.x.f.quant;
title_fig = {'Position X', 'Position Y', 'Velocity X', 'Velocity Y'};
for k=1:4
figure('name', 'SMC: Filtering estimates')
title(title_fig{k})
hold on
h = fill([1:t_max, t_max:-1:1], [x_f_quant{1}(k,:), fliplr(x_f_quant{2}(k,:))], 0);
set(h, 'edgecolor', 'none', 'facecolor', light_blue)
plot(x_f_mean(k, :), 'linewidth', 3)
plot(data.x_true(k,:), 'g')
xlabel('Time')
ylabel(title_fig{k})
legend({'95% credible interval', 'Filtering mean estimate', 'True value'},...
'location', 'Northwest')
legend boxoff
box off
end
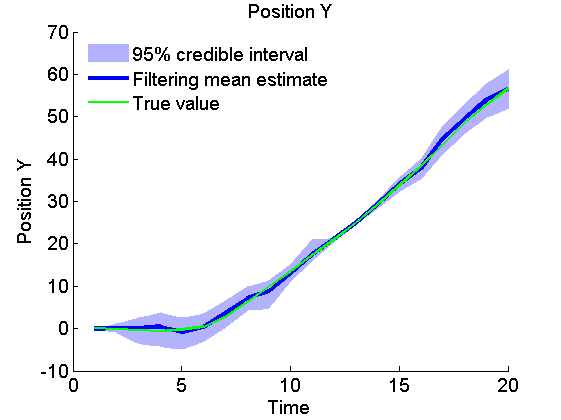
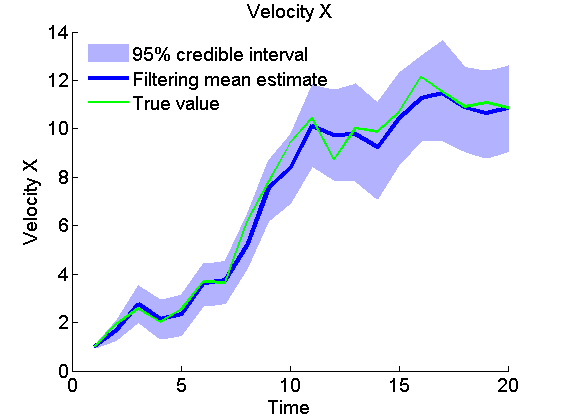
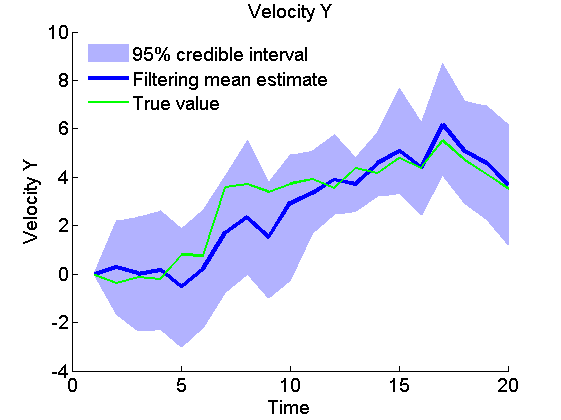
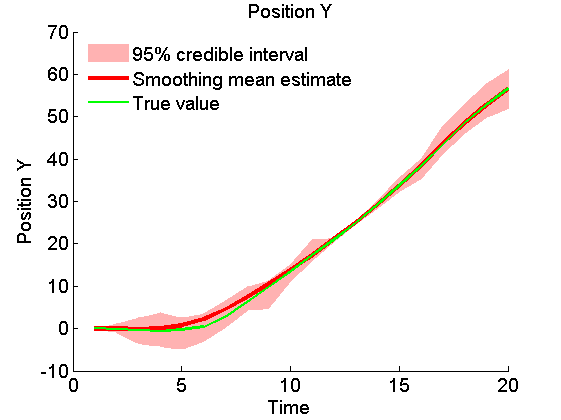
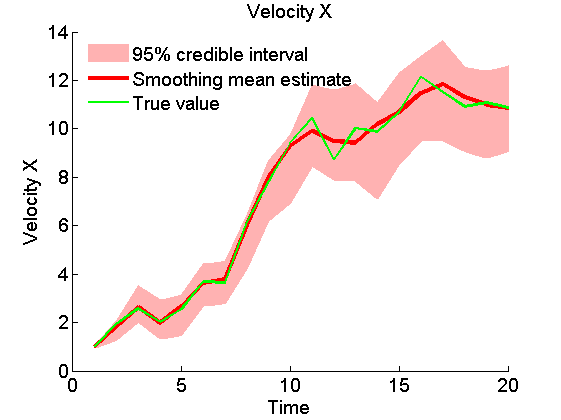
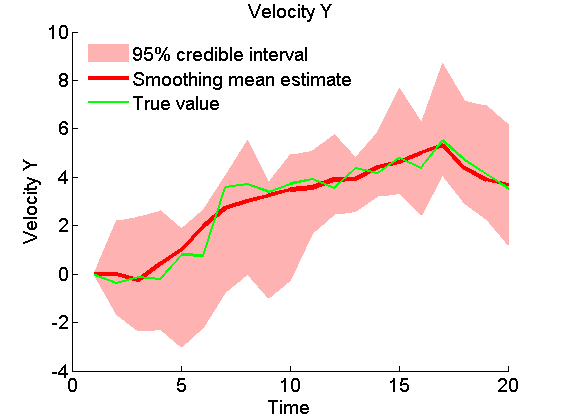
Plot Smoothing estimates
x_s_quant = summ_smc.x.s.quant; for k=1:4 figure('name', 'SMC: Smoothing estimates') title(title_fig{k}) hold on h = fill([1:t_max, t_max:-1:1], [x_f_quant{1}(k,:), fliplr(x_f_quant{2}(k,:))], 0); set(h, 'edgecolor', 'none', 'facecolor', light_red) plot(x_s_mean(k, :), 'r', 'linewidth', 3) plot(data.x_true(k,:), 'g') xlabel('Time') ylabel(title_fig{k}) legend({'95% credible interval', 'Smoothing mean estimate', 'True value'},... 'location', 'Northwest') legend boxoff box off end
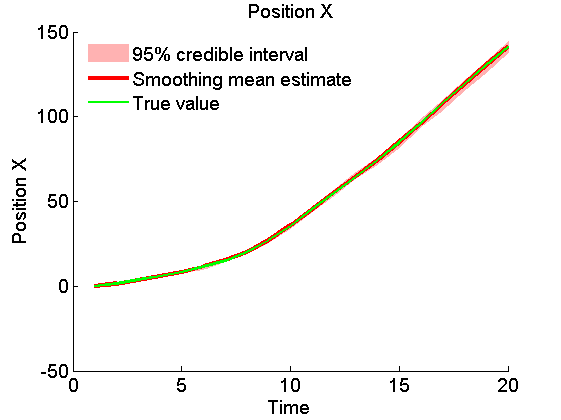
Clear model
biips_clear()